Young Researcher Paper Award 2025
🥇Winners
🥇Winners
Print: ISSN 0914-4935
Online: ISSN 2435-0869
Sensors and Materials
is an international peer-reviewed open access journal to provide a forum for researchers working in multidisciplinary fields of sensing technology.
Online: ISSN 2435-0869
Sensors and Materials
is an international peer-reviewed open access journal to provide a forum for researchers working in multidisciplinary fields of sensing technology.
Tweets by Journal_SandM
Sensors and Materials
is covered by Science Citation Index Expanded (Clarivate Analytics), Scopus (Elsevier), and other databases.
Instructions to authors
English 日本語
Instructions for manuscript preparation
English 日本語
Template
English
Publisher
MYU K.K.
Sensors and Materials
1-23-3-303 Sendagi,
Bunkyo-ku, Tokyo 113-0022, Japan
Tel: 81-3-3827-8549
Fax: 81-3-3827-8547
MYU Research, a scientific publisher, seeks a native English-speaking proofreader with a scientific background. B.Sc. or higher degree is desirable. In-office position; work hours negotiable. Call 03-3827-8549 for further information.

MYU Research
(proofreading and recording)

MYU K.K.
(translation service)

The Art of Writing Scientific Papers
(How to write scientific papers)
(Japanese Only)
is covered by Science Citation Index Expanded (Clarivate Analytics), Scopus (Elsevier), and other databases.
Instructions to authors
English 日本語
Instructions for manuscript preparation
English 日本語
Template
English
Publisher
MYU K.K.
Sensors and Materials
1-23-3-303 Sendagi,
Bunkyo-ku, Tokyo 113-0022, Japan
Tel: 81-3-3827-8549
Fax: 81-3-3827-8547
MYU Research, a scientific publisher, seeks a native English-speaking proofreader with a scientific background. B.Sc. or higher degree is desirable. In-office position; work hours negotiable. Call 03-3827-8549 for further information.

MYU Research
(proofreading and recording)

MYU K.K.
(translation service)

The Art of Writing Scientific Papers
(How to write scientific papers)
(Japanese Only)
Sensors and Materials, Volume 33, Number 1(2) (2021)
Copyright(C) MYU K.K.
Copyright(C) MYU K.K.
|
pp. 191-200
S&M2445 Perspective of Special Issue https://doi.org/10.18494/SAM.2021.3065 Published: January 22, 2021 Designer Interfaces for Label-free Biosensing [PDF] Tatsuro Goda (Received August 26, 2020; Accepted October 20, 2020) Keywords: aptamer, molecular switch, cell membrane mimicry, biomarker, influenza virus
Nowadays, there is huge demand for biosensing in point-of-care and wearable formats. Label-free biosensors meet the criteria because they are fast and straightforward without the need for labeling to detect target molecules. Instead, they have to detect a small amount of analyte among irrelevant molecules with a high signal-to-noise ratio by performing efficient biorecognition. Therefore, it is crucial to design and control the structure of the selective layer at the interface between the sample solution and the transducer. The selective layer contains capturing elements from natural or synthetic origins. Artificial receptors are a new class of components with improved stability, durability, and quality as compared with natural antibodies and enzymes. This review addresses the functionalization of the selective layer. It possesses smart, biomimetic, and non-fouling characteristics using synthetic receptors and molecules. Molecular switches made of an oligonucleotide can respond to the target upon environmental stimuli. Their responsiveness is tunable by designing the probe sequence. Cell membranes in which bioreceptors are embedded in self-assembled phospholipid bilayer membranes provide hints for developing an advanced biosensing platform. Cell membrane-mimetic surfaces are robust through covalent contacts and resist nonspecific adsorption. The adaptation of a cell surface receptor efficiently improves the binding specificity. These technologies are essential for developing next-generation label-free biosensors.
Corresponding author: Tatsuro Goda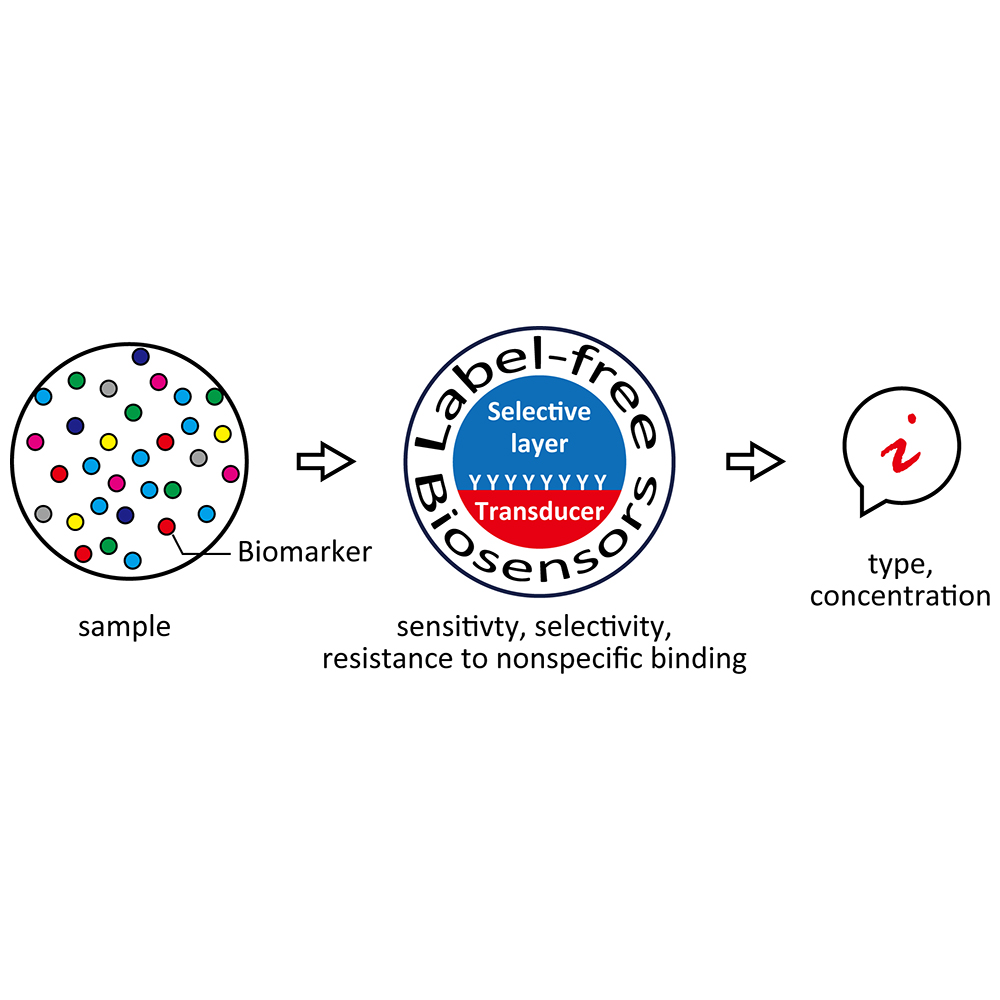  This work is licensed under a Creative Commons Attribution 4.0 International License. Cite this article Tatsuro Goda, Designer Interfaces for Label-free Biosensing, Sens. Mater., Vol. 33, No. 1, 2021, p. 191-200. |
Forthcoming Regular Issues
Forthcoming Special Issues
Special Issue on Novel Sensors, Materials, and Related Technologies on Artificial Intelligence of Things Applications
Guest editor, Teen-Hang Meen (National Formosa University), Wenbing Zhao (Cleveland State University), and Cheng-Fu Yang (National University of Kaohsiung)
Call for paper
Special Issue on Mobile Computing and Ubiquitous Networking for Smart Society
Guest editor, Akira Uchiyama (The University of Osaka) and Jaehoon Paul Jeong (Sungkyunkwan University)
Call for paper
Special Issue on Advanced Materials and Technologies for Sensor and Artificial- Intelligence-of-Things Applications (Selected Papers from ICASI 2026)
Guest editor, Sheng-Joue Young (National Yunlin University of Science and Technology)
Conference website
Call for paper
Special Issue on Innovations in Multimodal Sensing for Intelligent Devices, Systems, and Applications (submission closed)
Guest editor, Jiahui Yu (Research scientist, Zhejiang University), Kairu Li (Professor, Shenyang University of Technology), Yinfeng Fang (Professor, Hangzhou Dianzi University), Chin Wei Hong (Professor, Tokyo Metropolitan University), Zhiqiang Zhang (Professor, University of Leeds)
Call for paper
Special Issue on Multisource Sensors for Geographic Spatiotemporal Analysis and Social Sensing Technology Part 5
Guest editor, Prof. Bogang Yang (Beijing Institute of Surveying and Mapping) and Prof. Xiang Lei Liu (Beijing University of Civil Engineering and Architecture)
Special Issue on Advanced GeoAI for Smart Cities: Novel Data Modeling with Multi-source Sensor Data
Guest editor, Prof. Changfeng Jing (China University of Geosciences Beijing)
Call for paper
-
For more information of Special Issues (click here)
-
Special Issue on Materials, Devices, Circuits, and Analytical Methods for Various Sensors (Selected Papers from ICSEVEN 2025)
- Accepted papers (click here)
- Voltage Reflex and Equalization Charger for Series-connected Batteries
Cheng-Tao Tsai and Jia-Wei Lin
- Voltage Reflex and Equalization Charger for Series-connected Batteries
- Accepted papers (click here)
- Design and Development of a Fuzzy-logic-based Long-range Aquaculture System
Sheng-Tao Chen and Tai-I Chou
- Design and Development of a Fuzzy-logic-based Long-range Aquaculture System
Guest editor, Chien-Jung Huang (National University of Kaohsiung), Mu-Chun Wang (Minghsin University of Science and Technology), Shih-Hung Lin (Chung Shan Medical University), Ja-Hao Chen (Feng Chia University)
Conference website
Call for paper
Special Issue on Sensing and Data Analysis Technologies for Living Environment, Health Care, Production Management, and Engineering/Science Education Applications (2025)
Guest editor, Chien-Jung Huang (National University of Kaohsiung), Rey-Chue Hwang (I-Shou University), Ja-Hao Chen (Feng Chia University), Ba-Son Nguyen (Lac Hong University)
Call for paper
Special Issue on Advances in Sensors and Computational Intelligence for Industrial Applications
Guest editor, Chih-Hsien Hsia (National Ilan University)
Call for paper
Special Issue on AI-driven Sustainable Sensor Materials, Processes, and Circular Economy Applications
Guest editor, Shih-Chen Shi (National Cheng Kung University) and Tao-Hsing Chen (National Kaohsiung University of Science and Technology)
Call for paper
Special Issue on Intelligent Sensing and AI-driven Optimization for Sustainable Smart Manufacturing
Guest editor, Cheng-Chi Wang (National Sun Yat-sen University)
Call for paper
- Accepted papers (click here)
Copyright(C) MYU K.K. All Rights Reserved.
